

DNA fingerprinting is a way to identify using DNA. Some applications of DNA fingerprinting include:
DNA fingerprinting involves multiple biotechnologies, including PCR (see the chapter on PCR), but here this laboratory focuses on creating DNA fingerprints using restriction enzymes and visualizing the DNA fingerprints using gel electrophoresis.
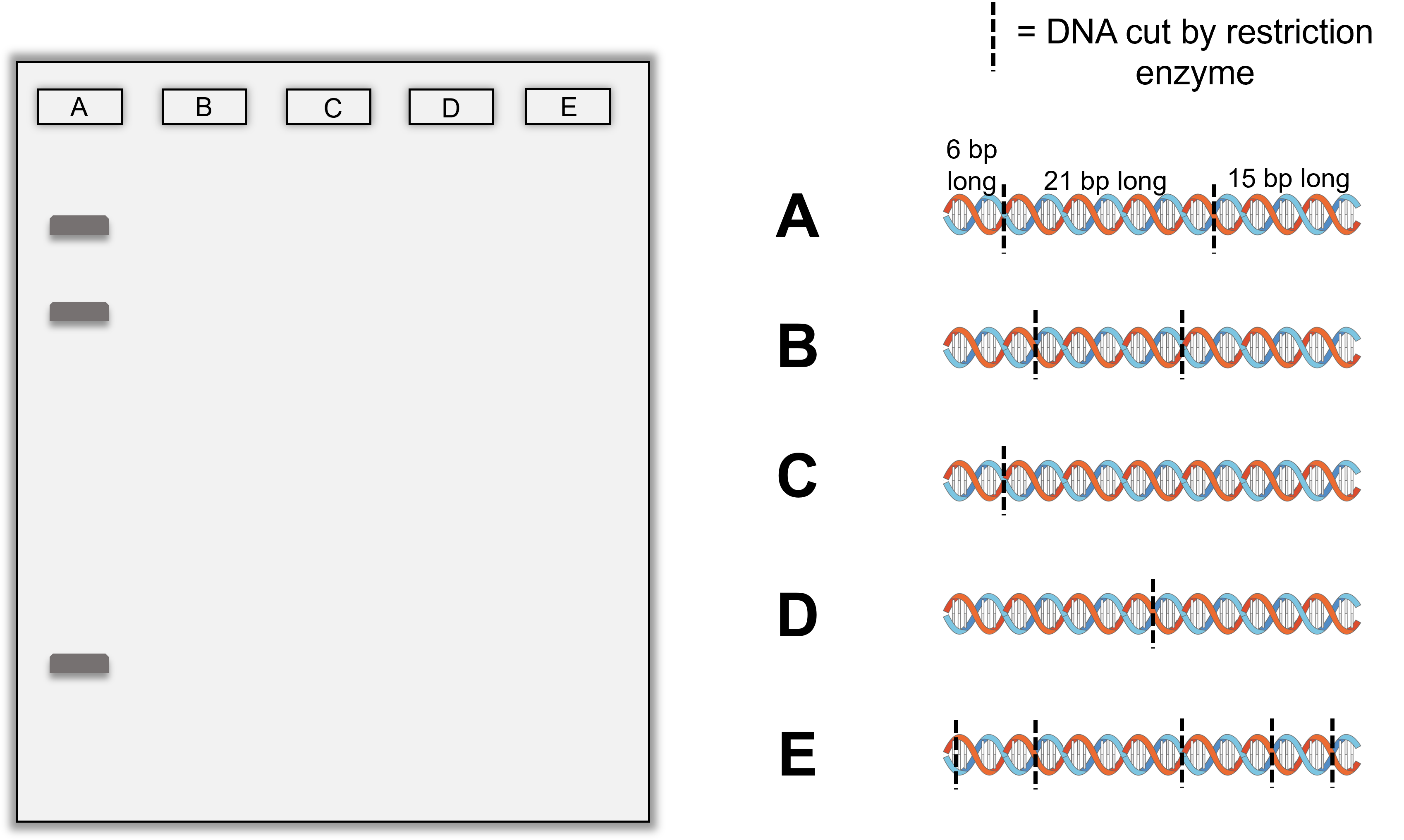
DNA fingerprints are created by first isolating DNA from an unknown sample to be identified and compared with known samples. If the samples match, it enables identification. The isolated DNA (i.e. DNA that has been removed from cells and other cell components) is mixed with a restriction enzyme to create a fingerprint. The restriction enzyme will cut the DNA in a pattern that will differ from DNA from other sources, unless the identify of the DNA is the same (matching known and unknown samples enables identification).
The DNA fragments produced by the restriction enzyme are separated by size using an approach called gel electrophoresis (see the Gel Electrophoresis section below). The result is a pattern of bands that can be compared with other patterns from known samples. If fingerprints match, it likely means that the DNA originated from the same organism. For paternity testing, half of the fingerprint will originate from the biological mother and half of the fingerprint will originate from the biological father.
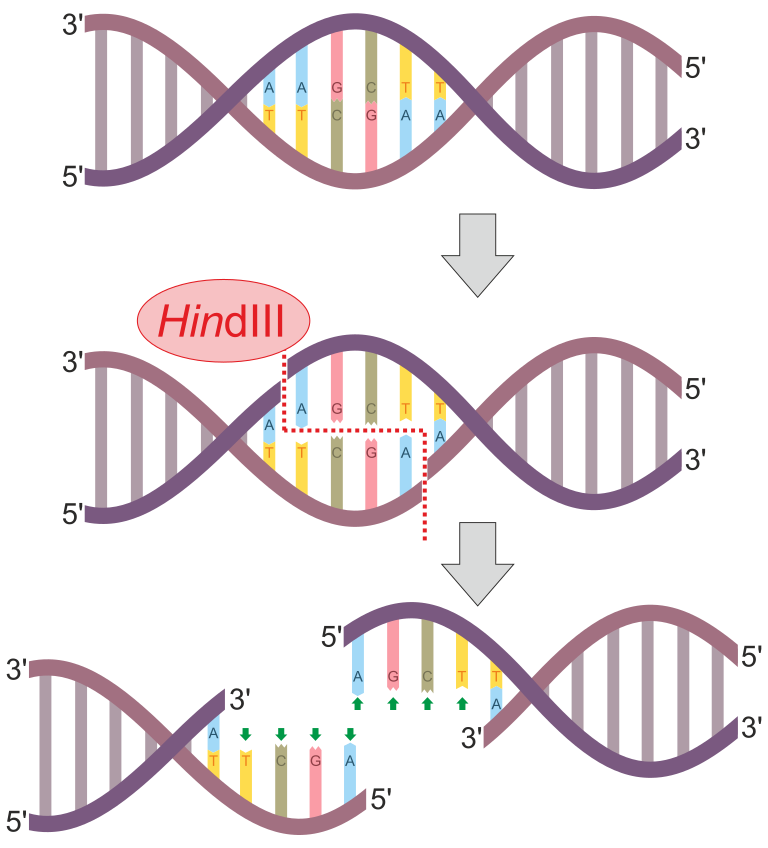
Restriction enzymes are found in some bacteria and have been isolated to use for a variety of biotechnologies such as DNA fingerprinting. These enzymes cut DNA at a characteristic recognition site. Recognition sites are different for each restriction enzyme. Typically, recognition sites are palindromic, that is they read the same backwards and forwards. Ordinary words that are palindromic include "mom," "dad," "wow," and "racecar." With DNA, a palindrome is based on reading one DNA strand 5' to 3' and comparing it with its complement DNA strand as read 5' to 3'. For example:
Notice that the complementary DNA strands above, if reading from the 5' end, have the same sequence: 5'-GAATTC-3'. This example is a recognition sequence from the restriction enzyme known as EcoRI. The "Eco" part of the enzyme name comes from the fact that this enzyme originates from Escherichia coli (E. coli). The rest of the restriction enzyme name comes from the strain of the organism (in this case the R strain) and the order in which the enzyme was discovered (in this case it is the first restriction enzyme discovered from the R strain of E. coli - I is the Roman numeral for 1).
In the example of EcoRI, this enzyme cuts the DNA between the "G" and the "A" on the 5' side. As a result, both the top and bottom DNA strands are cut and only held together by the few hydrogen bonds between the 5'-AATT-3' on both strands. Because of this (and that hydrogen bonds are rather weak attractions), the two DNA strands fully separate leaving the 5'-AATT-3' overhangs on both broken strands. These overhangs are called sticky ends.
Restriction enzymes will identify every location on a DNA molecule with the recognition sequence and cut the DNA there. This means the a restriction enzyme will likely make multiple cuts in the DNA. This will produce DNA fragments of different numbers of fragments with different sizes based on the base sequence of the DNA. The fragment sizes and number of fragments produces the DNA fingerprint used for identification purposes in DNA fingerprinting.
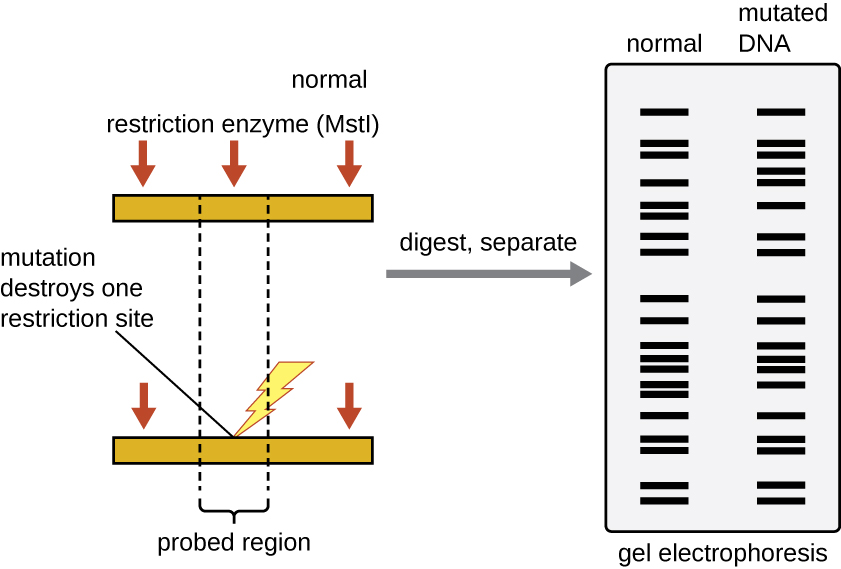
Restriction enzyme recognition sites are short (only a few nucleotides long), sequence-specific palindromes, and may be found throughout the genome. Thus, differences in DNA sequences in the genomes of individuals will lead to differences in distribution of restriction enzyme recognition sites that can be visualized as distinct fingerprints using a technique called gel electrophoresis (see the seciton on Gel Electrophoresis below). Restriction fragment length polymorphism (RFLP) analysis compares DNA banding patterns of different DNA samples after restriction digestion.
RFLP analysis has many practical applications in both medicine and forensic science. For example, epidemiologists use RFLP analysis to track and identify the source of specific microorganisms implicated in outbreaks of food poisoning or certain infectious diseases. RFLP analysis can also be used on human DNA to determine inheritance patterns of chromosomes with variant genes, including those associated with heritable diseases or to establish paternity.
Forensic scientists use RFLP analysis as a form of DNA fingerprinting, which is useful for analyzing DNA obtained from crime scenes, suspects, and victims. DNA samples are collected, the numbers of copies of the sample DNA molecules are increased using PCR, and then subjected to restriction enzyme digestion and agarose gel electrophoresis to generate specific banding patterns. By comparing the banding patterns of samples collected from the crime scene against those collected from suspects or victims, investigators can definitively determine whether DNA evidence collected at the scene was left behind by suspects or victims.
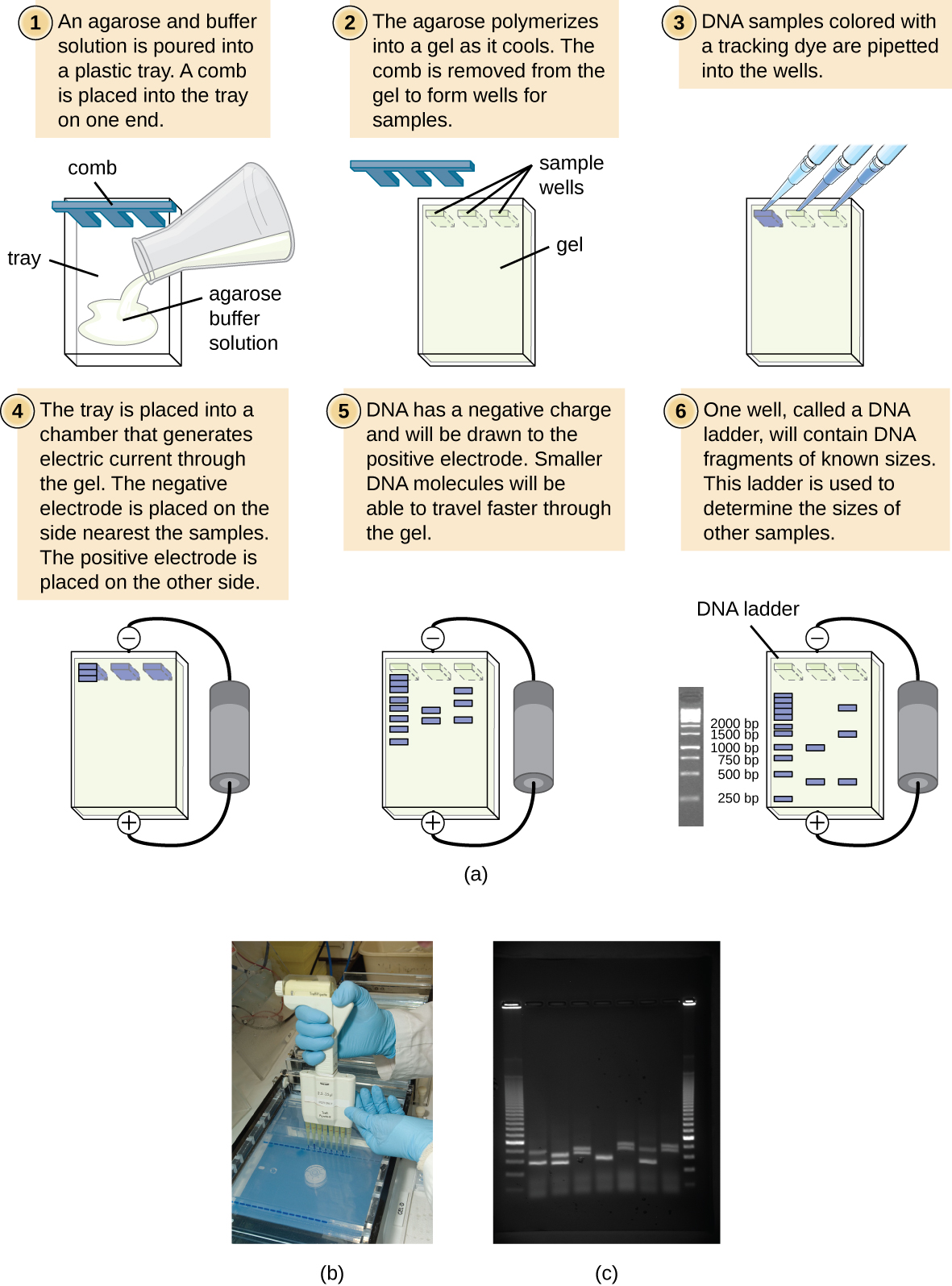
Gel electrophoresis is a technique commonly used to separate biological molecules based on size and biochemical characteristics, such as charge and polarity. Agarose gel electrophoresis is widely used to separate DNA (or RNA) of varying sizes that may be generated by restriction enzyme digestion (such as DNA fingerprinting / RFLP analysis) or by other means, such as the PCR.
DNA molecules have an overall negative charge. This is due to negative charges on the phosphate groups of its nucleotides. As a result, DNA can be pulled toward a positive charge. This is how gel electrophoresis pulls DNA through an agarose gel.
Due to its negatively charged backbone, DNA is strongly attracted to a positive electrode. In agarose gel electrophoresis, the gel is oriented horizontally in a buffer solution. Samples are loaded into sample wells on the side of the gel closest to the negative electrode, then drawn through the molecular sieve of the agarose matrix toward the positive electrode. The agarose matrix impedes the movement of larger molecules through the gel, whereas smaller molecules pass through more readily. Thus, the distance of migration is inversely correlated to the size of the DNA fragment, with smaller fragments traveling a longer distance through the gel. Sizes of DNA fragments within a sample can be estimated by comparison to fragments of known size in a DNA ladder also run on the same gel.
Small DNA fragments travel farther through the electrophoresis gel than larger DNA fragments.
You can think about this as being analagous to rocks in a river. Large boulders do not move very far, even if the current is swift, but small pebbles are capable of moving great distances in river's current. Similarly, large DNA cannot move well through the gel and will remain closer to the wells and smaller DNA can move easily through the gel and will move farther away from the wells.
You are an epidemiologist responsible for tracking outbreaks of potentially deadly disease and coordinating responses to help contain the spread of these diseases. It has been reported that a new viral infection is spreading in the Midwestern United States. This infection has symptoms very similar to two pervious viral outbreaks:
With this new outbreak in the Midwest, you believe that the virus causing the infections is either the a virus similar to the East Coast outbreak or the West Coast outbreak. How you organize the response to this outbreak is going to be dependent on whether the virus in the Midwest is the more severe strain or the less severe strain.
To determine if the virus is the East Coast virus or the West Coast virus, you conduct RFLP analysis (aka DNA fingerprinting) by extracting the viral DNA and digesting it with a restriction enzyme. To visualize the DNA fingerprint and compare it with the DNA fingerprint of the East Coast and West Coast viruses, you must use gel electrophoresis and then analyze the results.
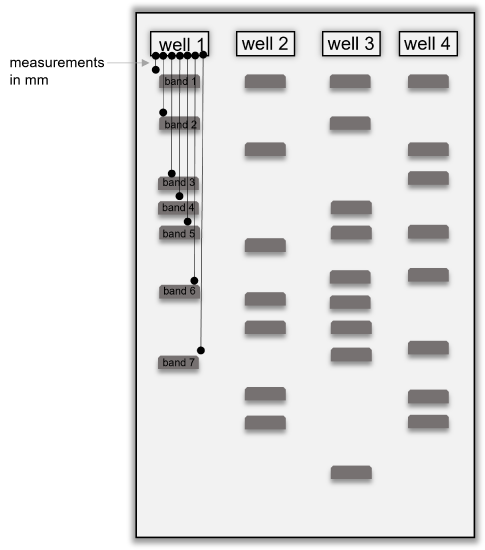
Table 1: Results from gel electrophoresis of DNA fingerprints of East coast virus, West coast virus, and Midwest virus. Some of the columns and rows may not be used in the table below, depending on how your gel was set up and the number of bands in each lane.
| well 1 | well 2 | well 3 | well 4 | well 5 | well 6 |
|---|---|---|---|---|---|
| sample in this well: | |||||
| band 1 migration (mm) | |||||
| band 2 migration (mm) | |||||
| band 3 migration (mm) | |||||
| band 4 migration (mm) | |||||
| band 5 migration (mm) | |||||
| band 6 migration (mm) | |||||
| band 7 migration (mm) | |||||
| band 8 migration (mm) | |||||
| band 9 migration (mm) | |||||
| band 10 migration (mm) |
This page titled 1.32: DNA Fingerprinting is shared under a CC BY-NC-SA 4.0 license and was authored, remixed, and/or curated by Rosanna Hartline.